Note
Go to the end to download the full example code
The Basics of Running a Meta-Analysis
Here we walk through the basic steps of running a meta-analysis with PyMARE.
Start with the necessary imports
from pprint import pprint
from pymare import core, datasets, estimators
Load the data
We will use the Michael et al.[1] dataset, which comes from the metadat library [2].
We only want to do a mean analysis, so we won’t have any covariates except for an intercept.
data, meta = datasets.michael2013()
dset = core.Dataset(data=data, y="yi", v="vi", X=None, add_intercept=True)
dset.to_df()
Now we fit a model
You must first initialize the estimator, after which you can use
fit() to fit the model to
numpy arrays, or
fit_dataset() to fit it to
a Dataset.
Tip
We generally recommend using
fit_dataset() over
fit().
There are a number of methods, such as
get_heterogeneity_stats() and
permutation_test(),
which only work when the Estimator is fitted to a Dataset.
However, fit() requires
less memory than fit_dataset(),
so it can be useful for large-scale meta-analyses,
such as neuroimaging image-based meta-analyses.
The summary() function
will return a MetaRegressionResults object,
which contains the results of the analysis.
est = estimators.WeightedLeastSquares().fit_dataset(dset)
results = est.summary()
results.to_df()
We can also extract some useful information from the results object
The get_heterogeneity_stats()
method will calculate heterogeneity statistics.
{'H': array([1.19590745]),
'I^2': array([30.07944649]),
'Q': array([15.73214091]),
'p(Q)': array([0.15136996])}
The get_re_stats() method will
estimate the confidence interval for 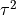 .
.
{'ci_l': array([0.]), 'ci_u': array([0.04013725]), 'tau^2': 0.0}
The permutation_test() method
will run a permutation test to estimate more accurate p-values.
perm_results = results.permutation_test(n_perm=1000)
perm_results.to_df()
References
Total running time of the script: (0 minutes 0.032 seconds)